biosppy.signals
This sub-package provides methods to process common physiological signals (biosignals).
Modules
biosppy.signals.abp
This module provides methods to process Arterial Blood Pressure (ABP) signals.
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.abp.abp(signal=None, sampling_rate=1000.0, show=True)[source]
Process a raw ABP signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw ABP signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered ABP signal.
onsets (array) – Indices of ABP pulse onsets.
heart_rate_ts (array) – Heart rate time axis reference (seconds).
heart_rate (array) – Instantaneous heart rate (bpm).
- biosppy.signals.abp.find_onsets_zong2003(signal=None, sampling_rate=1000.0, sm_size=None, size=None, alpha=2.0, wrange=None, d1_th=0, d2_th=None)[source]
Determine onsets of ABP pulses. Skips corrupted signal parts. Based on the approach by Zong et al. [Zong03]. :param signal: Input filtered ABP signal. :type signal: array :param sampling_rate: Sampling frequency (Hz). :type sampling_rate: int, float, optional :param sm_size: Size of smoother kernel (seconds).
Defaults to 0.25
- Parameters:
size (int, optional) – Window to search for maxima (seconds). Defaults to 5
alpha (float, optional) – Normalization parameter. Defaults to 2.0
wrange (int, optional) – The window in which to search for a peak (seconds). Defaults to 0.1
d1_th (int, optional) – Smallest allowed difference between maxima and minima. Defaults to 0
d2_th (int, optional) – Smallest allowed time between maxima and minima (seconds), Defaults to 0.15
- Returns:
onsets (array) – Indices of ABP pulse onsets.
References
[Zong03]W Zong, T Heldt, GB Moody and RG Mark, “An Open-source Algorithm to Detect Onset of Arterial Blood Pressure Pulses”, IEEE Comp. in Cardiology, vol. 30, pp. 259-262, 2003
biosppy.signals.acc
This module provides methods to process Acceleration (ACC) signals. Implemented code assumes ACC acquisition from a 3 orthogonal axis reference system.
- copyright:
2015-2023 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
Authors: Afonso Ferreira Diogo Vieira
- biosppy.signals.acc.acc(signal=None, sampling_rate=100.0, units=None, path=None, show=True, interactive=False)[source]
Process a raw ACC signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw ACC signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
interactive (bool, optional) – If True, shows an interactive plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
signal (array) – Raw (unfiltered) ACC signal.
vm (array) – Vector Magnitude feature of the signal.
sm (array) – Signal Magnitude feature of the signal.
freq_features (dict) – Positive Frequency domains (Hz) of the signal.
amp_features (dict) – Normalized Absolute Amplitudes of the signal.
- biosppy.signals.acc.activity_index(signal=None, sampling_rate=100.0, window_1=5, window_2=60)[source]
Compute the activity index of an ACC signal.
Follows the method described in [Lin18], the activity index is computed as follows:
Calculate the ACC magnitude if the signal is triaxial
2) Calculate the standard deviation of the ACC magnitude for each ‘window_1’ seconds 3) The activity index will be the mean standard deviation for each ‘window_2’ seconds
- Parameters:
signal (array) – Raw ACC signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
window_1 (int, float, optional) – Window length (seconds) for the first moving average filter. Default: 5
window_2 (int, float, optional) – Window length (seconds) for the second moving average filter. Default: 60
- Returns:
ts (array) – Time axis reference (seconds).
activity_index (array) – Activity index of the signal.
References
[Lin18]W.-Y. Lin, V. Verma, M.-Y. Lee, C.-S. Lai, Activity
Monitoring with a Wrist-Worn, Accelerometer-Based Device, Micromachines 9 (2018) 450
- biosppy.signals.acc.frequency_domain_feature_extractor(signal=None, sampling_rate=100.0)[source]
Extracts the FFT from each ACC sub-signal (x, y, z), given the signal itself.
- Parameters:
signal (array) – Input ACC signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
- Returns:
freq_features (dict) – Dictionary of positive frequencies (Hz) for all sub-signals.
amp_features (dict) – Dictionary of Normalized Absolute Amplitudes for all sub-signals.
- biosppy.signals.acc.time_domain_feature_extractor(signal=None)[source]
Extracts the vector magnitude and signal magnitude features from an input ACC signal, given the signal itself.
- Parameters:
signal (array) – Input ACC signal.
- Returns:
vm_features (array) – Extracted Vector Magnitude (VM) feature.
sm_features (array) – Extracted Signal Magnitude (SM) feature.
biosppy.signals.bvp
This module provides methods to process Blood Volume Pulse (BVP) signals.
——– DEPRECATED ——– PLEASE, USE THE PPG MODULE This module was left for compatibility —————————-
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.bvp.bvp(signal=None, sampling_rate=1000.0, path=None, show=True)[source]
Process a raw BVP signal and extract relevant signal features using default parameters. :param signal: Raw BVP signal. :type signal: array :param sampling_rate: Sampling frequency (Hz). :type sampling_rate: int, float, optional :param path: If provided, the plot will be saved to the specified file. :type path: str, optional :param show: If True, show a summary plot. :type show: bool, optional
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered BVP signal.
onsets (array) – Indices of BVP pulse onsets.
heart_rate_ts (array) – Heart rate time axis reference (seconds).
heart_rate (array) – Instantaneous heart rate (bpm).
biosppy.signals.ecg
This module provides methods to process Electrocardiographic (ECG) signals. Implemented code assumes a single-channel Lead I like ECG signal.
- copyright:
2015-2023 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.ecg.ASI_segmenter(signal=None, sampling_rate=1000.0, Pth=5.0)[source]
ECG R-peak segmentation algorithm.
- Parameters:
signal (array) – Input ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
Pth (int, float, optional) – Free parameter used in exponential decay
- Returns:
rpeaks (array) – R-peak location indices.
References
Modification by Tiago Rodrigues, based on: [R. Gutiérrez-rivas 2015] Novel Real-Time Low-Complexity QRS Complex Detector
Based on Adaptive Thresholding. Vol. 15,no. 10, pp. 6036–6043, 2015.
- [D. Sadhukhan] R-Peak Detection Algorithm for Ecg using Double Difference
And RRInterval Processing. Procedia Technology, vol. 4, pp. 873–877, 2012.
- biosppy.signals.ecg.ZZ2018(signal, detector_1, detector_2, fs=1000, search_window=100, nseg=1024, mode='simple')[source]
- biosppy.signals.ecg.bSQI(detector_1, detector_2, fs=1000.0, mode='simple', search_window=150)[source]
Comparison of the output of two detectors.
- Parameters:
detector_1 (array) – Output of the first detector.
detector_2 (array) – Output of the second detector.
fs (int, optional) – Sampling rate, in Hz.
mode (str, optional) – If ‘simple’, return only the percentage of beats detected by both. If ‘matching’, return the peak matching degree. If ‘n_double’ returns the number of matches divided by the sum of all minus the matches.
search_window (int, optional) – Search window around each peak, in ms.
- Returns:
bSQI (float) – Performance of both detectors.
- biosppy.signals.ecg.christov_segmenter(signal=None, sampling_rate=1000.0)[source]
ECG R-peak segmentation algorithm.
Follows the approach by Christov [Chri04].
- Parameters:
signal (array) – Input filtered ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
- Returns:
rpeaks (array) – R-peak location indices.
References
[Chri04]Ivaylo I. Christov, “Real time electrocardiogram QRS detection using combined adaptive threshold”, BioMedical Engineering OnLine 2004, vol. 3:28, 2004
- biosppy.signals.ecg.compare_segmentation(reference=None, test=None, sampling_rate=1000.0, offset=0, minRR=None, tol=0.05)[source]
Compare the segmentation performance of a list of R-peak positions against a reference list.
- Parameters:
reference (array) – Reference R-peak location indices.
test (array) – Test R-peak location indices.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
offset (int, optional) – Constant a priori offset (number of samples) between reference and test R-peak locations.
minRR (float, optional) – Minimum admissible RR interval (seconds).
tol (float, optional) – Tolerance between corresponding reference and test R-peak locations (seconds).
- Returns:
TP (int) – Number of true positive R-peaks.
FP (int) – Number of false positive R-peaks.
performance (float) – Test performance; TP / len(reference).
acc (float) – Accuracy rate; TP / (TP + FP).
err (float) – Error rate; FP / (TP + FP).
match (list) – Indices of the elements of ‘test’ that match to an R-peak from ‘reference’.
deviation (array) – Absolute errors of the matched R-peaks (seconds).
mean_deviation (float) – Mean error (seconds).
std_deviation (float) – Standard deviation of error (seconds).
mean_ref_ibi (float) – Mean of the reference interbeat intervals (seconds).
std_ref_ibi (float) – Standard deviation of the reference interbeat intervals (seconds).
mean_test_ibi (float) – Mean of the test interbeat intervals (seconds).
std_test_ibi (float) – Standard deviation of the test interbeat intervals (seconds).
- biosppy.signals.ecg.correct_rpeaks(signal=None, rpeaks=None, sampling_rate=1000.0, tol=0.05)[source]
Correct R-peak locations to the maximum within a tolerance.
- Parameters:
signal (array) – ECG signal.
rpeaks (array) – R-peak location indices.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
tol (int, float, optional) – Correction tolerance (seconds).
- Returns:
rpeaks (array) – Cerrected R-peak location indices.
Notes
The tolerance is defined as the time interval
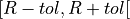 .
.
- biosppy.signals.ecg.ecg(signal=None, sampling_rate=1000.0, units=None, path=None, show=True, interactive=False)[source]
Process a raw ECG signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
interactive (bool, optional) – If True, shows an interactive plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered ECG signal.
rpeaks (array) – R-peak location indices.
templates_ts (array) – Templates time axis reference (seconds).
templates (array) – Extracted heartbeat templates.
heart_rate_ts (array) – Heart rate time axis reference (seconds).
heart_rate (array) – Instantaneous heart rate (bpm).
- biosppy.signals.ecg.engzee_segmenter(signal=None, sampling_rate=1000.0, threshold=0.48)[source]
ECG R-peak segmentation algorithm.
Follows the approach by Engelse and Zeelenberg [EnZe79] with the modifications by Lourenco et al. [LSLL12].
- Parameters:
signal (array) – Input filtered ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
threshold (float, optional) – Detection threshold.
- Returns:
rpeaks (array) – R-peak location indices.
References
[EnZe79]W. Engelse and C. Zeelenberg, “A single scan algorithm for QRS detection and feature extraction”, IEEE Comp. in Cardiology, vol. 6, pp. 37-42, 1979
[LSLL12]A. Lourenco, H. Silva, P. Leite, R. Lourenco and A. Fred, “Real Time Electrocardiogram Segmentation for Finger Based ECG Biometrics”, BIOSIGNALS 2012, pp. 49-54, 2012
- biosppy.signals.ecg.extract_heartbeats(signal=None, rpeaks=None, sampling_rate=1000.0, before=0.2, after=0.4)[source]
Extract heartbeat templates from an ECG signal, given a list of R-peak locations.
- Parameters:
signal (array) – Input ECG signal.
rpeaks (array) – R-peak location indices.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
before (float, optional) – Window size to include before the R peak (seconds).
after (int, optional) – Window size to include after the R peak (seconds).
- Returns:
templates (array) – Extracted heartbeat templates.
rpeaks (array) – Corresponding R-peak location indices of the extracted heartbeat templates.
- biosppy.signals.ecg.fSQI(ecg_signal, fs=1000.0, nseg=1024, num_spectrum=[5, 20], dem_spectrum=None, mode='simple')[source]
Returns the ration between two frequency power bands.
- Parameters:
ecg_signal (array) – ECG signal.
fs (float, optional) – ECG sampling frequency, in Hz.
nseg (int, optional) – Frequency axis resolution.
num_spectrum (array, optional) – Frequency bandwidth for the ratio’s numerator, in Hz.
dem_spectrum (array, optional) – Frequency bandwidth for the ratio’s denominator, in Hz. If None, then the whole spectrum is used.
mode (str, optional) – If ‘simple’ just do the ration, if is ‘bas’, then do 1 - num_power.
- Returns:
Ratio (float) – Ratio between two powerbands.
- biosppy.signals.ecg.gamboa_segmenter(signal=None, sampling_rate=1000.0, tol=0.002)[source]
ECG R-peak segmentation algorithm.
Follows the approach by Gamboa.
- Parameters:
signal (array) – Input filtered ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
tol (float, optional) – Tolerance parameter.
- Returns:
rpeaks (array) – R-peak location indices.
- biosppy.signals.ecg.getPPositions(ecg_proc=None, show=False)[source]
- Different ECG Waves (Q, R, S, …) are not present or are not so clear to identify in all ECG signals (I II III V1 V2 V3, …)
For P wave we suggest to use signals II, V1, aVF . Avoid I, III, V1, V2, V3, V4, V5, AVL
- Parameters:
signal (object) –
ecg. (object return by the function) –
show (bool, optional) –
True (If) –
sample/template. (show a plot of the P Positions on every signal) –
- Returns:
P_positions (array) – Array with all P positions on the signal
P_start_ positions (array) – Array with all P start positions on the signal
P_end_ positions (array) – Array with all P end positions on the signal
- biosppy.signals.ecg.getQPositions(ecg_proc=None, show=False)[source]
Different ECG Waves (Q, R, S, …) are not present or are not so clear to identify in all ECG signals (I II III V1 V2 V3, …) For Q wave we suggest to use signals I, aVL . Avoid II, III, V1, V2, V3, V4, aVR, aVF
- Parameters:
signal (object) –
ecg. (object return by the function) –
show (bool, optional) –
True (If) –
sample/template. (show a plot of the Q Positions on every signal) –
- Returns:
Q_positions (array) – Array with all Q positions on the signal
Q_start_ positions (array) – Array with all Q start positions on the signal
- biosppy.signals.ecg.getSPositions(ecg_proc=None, show=False)[source]
- Different ECG Waves (Q, R, S, …) are not present or are not so clear to identify in all ECG signals (I II III V1 V2 V3, …)
For S wave we suggest to use signals V1, V2, V3. Avoid I, V5, V6, aVR, aVL
- Parameters:
signal (object) –
ecg. (object return by the function) –
show (bool, optional) –
True (If) –
sample/template. (show a plot of the S Positions on every signal) –
- Returns:
S_positions (array) – Array with all S positions on the signal
S_end_ positions (array) – Array with all S end positions on the signal
- biosppy.signals.ecg.getTPositions(ecg_proc=None, show=False)[source]
Different ECG Waves (Q, R, S, …) are not present or are not so clear to identify in all ECG signals (I II III V1 V2 V3, …) For T wave we suggest to use signals V4, v5 (II, V3 have good results, but in less accuracy) . Avoid I, V1, V2, aVR, aVL
- Parameters:
signal (object) –
ecg. (object return by the function) –
show (bool, optional) –
True (If) –
sample/template. (show a plot of the T Positions on every signal) –
- Returns:
T_positions (array) – Array with all T positions on the signal
T_start_ positions (array) – Array with all T start positions on the signal
T_end_ positions (array) – Array with all T end positions on the signal
- biosppy.signals.ecg.hamilton_segmenter(signal=None, sampling_rate=1000.0)[source]
ECG R-peak segmentation algorithm.
Follows the approach by Hamilton [Hami02].
- Parameters:
signal (array) – Input filtered ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
- Returns:
rpeaks (array) – R-peak location indices.
References
[Hami02]P.S. Hamilton, “Open Source ECG Analysis Software Documentation”, E.P.Limited, 2002
- biosppy.signals.ecg.kSQI(signal, fisher=True)[source]
Return the kurtosis of the signal
- Parameters:
signal (array) – ECG signal.
fisher (bool, optional) – If True,Fisher’s definition is used (normal ==> 0.0). If False, Pearson’s definition is used (normal ==> 3.0).
- Returns:
kurtosis (float) – Kurtosis value.
- biosppy.signals.ecg.pSQI(signal, f_thr=0.01)[source]
Return the flatline percentage of the signal
- Parameters:
signal (array) – ECG signal.
f_thr (float, optional) – Flatline threshold, in mV / sample
- Returns:
flatline_percentage (float) – Percentage of signal where the absolute value of the derivative is lower then the threshold.
- biosppy.signals.ecg.sSQI(signal)[source]
Return the skewness of the signal
- Parameters:
signal (array) – ECG signal.
- Returns:
skewness (float) – Skewness value.
- biosppy.signals.ecg.ssf_segmenter(signal=None, sampling_rate=1000.0, threshold=20, before=0.03, after=0.01)[source]
ECG R-peak segmentation based on the Slope Sum Function (SSF).
- Parameters:
signal (array) – Input filtered ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
threshold (float, optional) – SSF threshold.
before (float, optional) – Search window size before R-peak candidate (seconds).
after (float, optional) – Search window size after R-peak candidate (seconds).
- Returns:
rpeaks (array) – R-peak location indices.
biosppy.signals.eda
This module provides methods to process Electrodermal Activity (EDA) signals, also known as Galvanic Skin Response (GSR).
- copyright:
2015-2023 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.eda.basic_scr(signal=None)[source]
Basic method to extract Skin Conductivity Responses (SCR) from an EDA signal.
Follows the approach in [Gamb08].
- Parameters:
signal (array) – Input filtered EDA signal.
- Returns:
onsets (array) – Indices of the SCR onsets.
peaks (array) – Indices of the SCR peaks.
amplitudes (array) – SCR pulse amplitudes.
References
[Gamb08]Hugo Gamboa, “Multi-modal Behavioral Biometrics Based on HCI and Electrophysiology”, PhD thesis, Instituto Superior T{‘e}cnico, 2008
- biosppy.signals.eda.biosppy_decomposition(signal=None, sampling_rate=1000.0, method='smoother', onsets=None, **kwargs)[source]
Extracts EDL and EDR signals using either a smoothing filter or onsets’ interpolation.
- Parameters:
signal (array) – Input filtered EDA signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
method (str, optional) – Method to compute the edl signal: “smoother” to compute a smoothing filter; “onsets” to obtain edl by onsets’ interpolation.
onsets (array, optional) – List of onsets for the interpolation method.
kwargs (dict, optional) – window_size : Size of the smoother kernel (seconds).
- Returns:
edl (array) – Electrodermal level (EDL) signal.
edr (array) – Electrodermal response (EDR) signal.
References
[KiBK04]K.H. Kim, S.W. Bang, and S.R. Kim, “Emotion recognition system
using short-term monitoring of physiological signals”, Med. Biol. Eng. Comput., vol. 42, pp. 419-427, 2004
- biosppy.signals.eda.cvx_decomposition(signal=None, sampling_rate=1000.0, tau0=2.0, tau1=0.7, delta_knot=10.0, alpha=0.0008, gamma=0.01, solver=None, options={'reltol': 1e-09})[source]
Performs EDA decomposition using the cvxEDA algorithm.
This function was originally developed by Luca Citi and Alberto Greco. You can find the original code and repository at: https://github.com/lciti/cvxEDA
If you use this function in your work, please cite the original authors as follows: A Greco, G Valenza, A Lanata, EP Scilingo, and L Citi “cvxEDA: a Convex Optimization Approach to Electrodermal Activity Processing” IEEE Transactions on Biomedical Engineering, 2015.
This function is used under the terms of the GNU General Public License v3.0 (GPLv3). You should comply with the GPLv3 if you use this code (see ‘License’ section below).
Copyright (C) 2014-2015 Luca Citi, Alberto Greco
- Parameters:
signal (array) – Observed EDA signal (we recommend normalizing it: y = zscore(y))
sampling_rate (int, float) – Sampling frequency (Hz).
tau0 (float) – Slow time constant of the Bateman function
tau1 (float) – Fast time constant of the Bateman function
delta_knot (float) – Time between knots of the tonic spline function
alpha (float) – Penalization for the sparse SMNA driver
gamma (float) – Penalization for the tonic spline coefficients
solver (ndarray) – Sparse QP solver to be used, see cvxopt.solvers.qp
options (dict) – solver options, see: http://cvxopt.org/userguide/coneprog.html#algorithm-parameters
- Returns:
edr (array) – Phasic component
smna (array) – Sparse SMNA driver of phasic component
edl (array) – Tonic component
tonic_coeff (array) – Coefficients of tonic spline
linear_drift (array) – Offset and slope of the linear drift term
res (array) – Model residuals
obj (array) – Value of objective function being minimized (eq 15 of paper)
References
[cvxEDA]A Greco, G Valenza, A Lanata, EP Scilingo, and L Citi
“cvxEDA: a Convex Optimization Approach to Electrodermal Activity Processing” IEEE Transactions on Biomedical Engineering, 2015. DOI: 10.1109/TBME.2015.2474131
[Figner2011]Figner, Bernd & Murphy, Ryan. (2011). Using skin
conductance in judgment and decision making research. A Handbook of Process Tracing Methods for Decision Research.
License
The cvxEDA function is distributed under the GNU General Public License v3.0 (GPLv3). For details, please see the full license text at: https://www.gnu.org/licenses/gpl-3.0.en.html
This code is provided as-is, without any warranty or support from the original authors.
Notes
Changes from original code: - ‘y’ -> ‘signal’ - ‘delta’ -> 1. / ‘sampling_rate’
- biosppy.signals.eda.eda(signal=None, sampling_rate=1000.0, units=None, path=None, show=True)[source]
Process a raw EDA signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw EDA signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered EDA signal.
edr (array) – Electrodermal response (EDR) signal.
edl (array) – Electrodermal level (EDL) signal.
onsets (array) – Indices of SCR pulse onsets.
peaks (array) – Indices of the SCR peaks.
amplitudes (array) – SCR pulse amplitudes.
- biosppy.signals.eda.eda_events(signal=None, sampling_rate=1000.0, method='emotiphai', **kwargs)[source]
Returns characteristic EDA events.
- Parameters:
signal (array) – Input signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
method (str, optional) – Method to compute eda events: ‘emotiphai’, ‘kbk’ or ‘basic’.
kwargs (dict, optional) – Method parameters.
- Returns:
onsets (array) – Signal EDR events onsets.
peaks (array) – Signal EDR events peaks.
amps (array) – Signal EDR events Amplitudes.
phasic_rate (array) – Signal EDR events rate in 60s.
rise_times (array) – Rise times, i.e. onset-peak time difference.
half_rec (array) – Half Recovery times, i.e. time between peak and 63% amplitude.
six_rec (array) – 63 % recovery times, i.e. time between peak and 50% amplitude.
- biosppy.signals.eda.emotiphai_eda(signal=None, sampling_rate=1000.0, min_amplitude=0.1, filt=True, size=1.0)[source]
Returns characteristic EDA events.
- Parameters:
signal (array) – Input signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
min_amplitude (float, optional) – Minimum threshold by which to exclude SCRs.
filt (bool, optional) – Whether to filter signal to remove noise and low amplitude events.
size (float) – Size of the filter in seconds
- Returns:
onsets (array) – Indices of the SCR onsets.
peaks (array) – Indices of the SCR peaks.
amplitudes (array) – SCR pulse amplitudes.
- biosppy.signals.eda.kbk_scr(signal=None, sampling_rate=1000.0, min_amplitude=0.1)[source]
KBK method to extract Skin Conductivity Responses (SCR) from an EDA signal.
Follows the approach by Kim et al. [KiBK04].
- Parameters:
signal (array) – Input filtered EDA signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
min_amplitude (float, optional) – Minimum threshold by which to exclude SCRs.
- Returns:
onsets (array) – Indices of the SCR onsets.
peaks (array) – Indices of the SCR peaks.
amplitudes (array) – SCR pulse amplitudes.
References
[KiBK04]K.H. Kim, S.W. Bang, and S.R. Kim, “Emotion recognition system using short-term monitoring of physiological signals”, Med. Biol. Eng. Comput., vol. 42, pp. 419-427, 2004
- biosppy.signals.eda.rec_times(signal=None, sampling_rate=1000.0, onsets=None, peaks=None)[source]
Returns EDA recovery times.
- Parameters:
signal (array) – Input signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
onsets (array) – Indices of the SCR onsets.
peaks (array) – Indices of the SCR peaks.
- Returns:
half_rec (list) – Half Recovery times, i.e. time between peak and 50% amplitude.
six_rec (list) – 63 % recovery times, i.e. time between peak and 63% amplitude.
biosppy.signals.eeg
This module provides methods to process Electroencephalographic (EEG) signals.
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.eeg.car_reference(signal=None)[source]
Change signal reference to the Common Average Reference (CAR).
- Parameters:
signal (array) – Input EEG signal matrix; each column is one EEG channel.
- Returns:
signal (array) – Re-referenced EEG signal matrix; each column is one EEG channel.
- biosppy.signals.eeg.eeg(signal=None, sampling_rate=1000.0, labels=None, path=None, show=True)[source]
Process raw EEG signals and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw EEG signal matrix; each column is one EEG channel.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
labels (list, optional) – Channel labels.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered EEG signal.
features_ts (array) – Features time axis reference (seconds).
theta (array) – Average power in the 4 to 8 Hz frequency band; each column is one EEG channel.
alpha_low (array) – Average power in the 8 to 10 Hz frequency band; each column is one EEG channel.
alpha_high (array) – Average power in the 10 to 13 Hz frequency band; each column is one EEG channel.
beta (array) – Average power in the 13 to 25 Hz frequency band; each column is one EEG channel.
gamma (array) – Average power in the 25 to 40 Hz frequency band; each column is one EEG channel.
plf_pairs (list) – PLF pair indices.
plf (array) – PLF matrix; each column is a channel pair.
- biosppy.signals.eeg.get_plf_features(signal=None, sampling_rate=1000.0, size=0.25, overlap=0.5)[source]
Extract Phase-Locking Factor (PLF) features from EEG signals between all channel pairs.
- Parameters:
signal (array) – Filtered EEG signal matrix; each column is one EEG channel.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (float, optional) – Window size (seconds).
overlap (float, optional) – Window overlap (0 to 1).
- Returns:
ts (array) – Features time axis reference (seconds).
plf_pairs (list) – PLF pair indices.
plf (array) – PLF matrix; each column is a channel pair.
- biosppy.signals.eeg.get_power_features(signal=None, sampling_rate=1000.0, size=0.25, overlap=0.5)[source]
Extract band power features from EEG signals.
Computes the average signal power, with overlapping windows, in typical EEG frequency bands: * Theta: from 4 to 8 Hz, * Lower Alpha: from 8 to 10 Hz, * Higher Alpha: from 10 to 13 Hz, * Beta: from 13 to 25 Hz, * Gamma: from 25 to 40 Hz.
- Parameters:
array (signal) – Filtered EEG signal matrix; each column is one EEG channel.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (float, optional) – Window size (seconds).
overlap (float, optional) – Window overlap (0 to 1).
- Returns:
ts (array) – Features time axis reference (seconds).
theta (array) – Average power in the 4 to 8 Hz frequency band; each column is one EEG channel.
alpha_low (array) – Average power in the 8 to 10 Hz frequency band; each column is one EEG channel.
alpha_high (array) – Average power in the 10 to 13 Hz frequency band; each column is one EEG channel.
beta (array) – Average power in the 13 to 25 Hz frequency band; each column is one EEG channel.
gamma (array) – Average power in the 25 to 40 Hz frequency band; each column is one EEG channel.
biosppy.signals.emg
This module provides methods to process Electromyographic (EMG) signals.
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.emg.abbink_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, alarm_size=None, threshold=None, transition_threshold=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Abbink et al.. [Abb98].
- Parameters:
signal (array) – Input filtered EMG signal.
rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (int) – Detection window size (seconds).
alarm_size (int) – Number of amplitudes searched in the calculation of the transition index.
threshold (int, float) – Detection threshold.
transition_threshold (int, float) – Threshold used in the calculation of the transition index.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[Abb98]Abbink JH, van der Bilt A, van der Glas HW, “Detection of onset and termination of muscle activity in surface electromyograms”, Journal of Oral Rehabilitation, vol. 25, pp. 365–369, 1998
- biosppy.signals.emg.bonato_onset_detector(signal=None, rest=None, sampling_rate=1000.0, threshold=None, active_state_duration=None, samples_above_fail=None, fail_size=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Bonato et al. [Bo98].
- Parameters:
signal (array) – Input filtered EMG signal.
rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
threshold (int, float) – Detection threshold.
active_state_duration (int) – Minimum duration of the active state.
samples_above_fail (int) – Number of samples above the threshold level in a group of successive samples.
fail_size (int) – Number of successive samples.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[Bo98]Bonato P, D’Alessio T, Knaflitz M, “A statistical method for the measurement of muscle activation intervals from surface myoelectric signal during gait”, IEEE Transactions on Biomedical Engineering, vol. 45:3, pp. 287–299, 1998
- biosppy.signals.emg.emg(signal=None, sampling_rate=1000.0, units=None, path=None, show=True)[source]
Process a raw EMG signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw EMG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered EMG signal.
onsets (array) – Indices of EMG pulse onsets.
- biosppy.signals.emg.find_onsets(signal=None, sampling_rate=1000.0, size=0.05, threshold=None)[source]
Determine onsets of EMG pulses.
Skips corrupted signal parts.
- Parameters:
signal (array) – Input filtered EMG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (float, optional) – Detection window size (seconds).
threshold (float, optional) – Detection threshold.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
- biosppy.signals.emg.hodges_bui_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, threshold=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Hodges and Bui [HoBu96].
- Parameters:
signal (array) – Input filtered EMG signal.
rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (int) – Detection window size (seconds).
threshold (int, float) – Detection threshold.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[HoBu96]Hodges PW, Bui BH, “A comparison of computer-based methods for the determination of onset of muscle contraction using electromyography”, Electroencephalography and Clinical Neurophysiology - Electromyography and Motor Control, vol. 101:6, pp. 511-519, 1996
- biosppy.signals.emg.lidierth_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, threshold=None, active_state_duration=None, fail_size=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Lidierth. [Li86].
- Parameters:
signal (array) – Input filtered EMG signal.
rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (int) – Detection window size (seconds).
threshold (int, float) – Detection threshold.
active_state_duration (int) – Minimum duration of the active state.
fail_size (int) – Number of successive samples.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[Li86]Lidierth M, “A computer based method for automated measurement of the periods of muscular activity from an EMG and its application to locomotor EMGs”, ElectroencephClin Neurophysiol, vol. 64:4, pp. 378–380, 1986
- biosppy.signals.emg.londral_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, threshold=None, active_state_duration=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Londral et al. [Lon13].
- Parameters:
signal (array) – Input filtered EMG signal.
rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (int) – Detection window size (seconds).
threshold (int, float) – Scale factor for calculating the detection threshold.
active_state_duration (int) – Minimum duration of the active state.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[Lon13]Londral A, Silva H, Nunes N, Carvalho M, Azevedo L, “A wireless user-computer interface to explore various sources of biosignals and visual biofeedback for severe motor impairment”, Journal of Accessibility and Design for All, vol. 3:2, pp. 118–134, 2013
- biosppy.signals.emg.silva_onset_detector(signal=None, sampling_rate=1000.0, size=None, threshold_size=None, threshold=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Silva et al. [Sil12].
- Parameters:
signal (array) – Input filtered EMG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (int) – Detection window size (seconds).
threshold_size (int) – Window size for calculation of the adaptive threshold; must be bigger than the detection window size.
threshold (int, float) – Fixed threshold for the double criteria.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[Sil12]Silva H, Scherer R, Sousa J, Londral A , “Towards improving the usability of electromyographic interfacess”, Journal of Oral Rehabilitation, pp. 1–2, 2012
- biosppy.signals.emg.solnik_onset_detector(signal=None, rest=None, sampling_rate=1000.0, threshold=None, active_state_duration=None)[source]
Determine onsets of EMG pulses.
Follows the approach by Solnik et al. [Sol10].
- Parameters:
signal (array) – Input filtered EMG signal.
rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
threshold (int, float) – Scale factor for calculating the detection threshold.
active_state_duration (int) – Minimum duration of the active state.
- Returns:
onsets (array) – Indices of EMG pulse onsets.
processed (array) – Processed EMG signal.
References
[Sol10]Solnik S, Rider P, Steinweg K, DeVita P, Hortobágyi T, “Teager-Kaiser energy operator signal conditioning improves EMG onset detection”, European Journal of Applied Physiology, vol 110:3, pp. 489-498, 2010
biosppy.signals.pcg
This module provides methods to process Phonocardiography (PCG) signals.
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.pcg.ecg_based_segmentation(pcg_signal=None, ecg_signal=None, sampling_rate=1000.0, show=False)[source]
Assign state labels to PCG recording based on markers from simultaneous ECG signal.
Adapted to Python from original MATLAB code written by David Springer, 2016 (C), for comparison purposes in the paper [Springer15]. Available at: https://github.com/davidspringer/Springer-Segmentation-Code
Heart sounds timing durations were obtained from [Schimdt10].
- Parameters:
pcg_signal (array) – PCG signal to be segmented.
ecg_signal (array) – Simultaneous ECG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz) of both signals.
show (bool, optional) – If True, show a plot with the segmented signal.
- Returns:
states (array) – State labels of PCG recording
References
[Springer15]D.Springer, “Logistic Regression-HSMM-based Heart Sound Segmentation”, IEEE Trans. Biomed. Eng., In Press, 2015.
[Schimdt10]S. E. Schmidt et al., “Segmentation of heart sound recordings by a duration-dependent hidden Markov model”, Physiol. Meas., 2010
- biosppy.signals.pcg.find_peaks(signal=None, sampling_rate=1000.0)[source]
Finds the peaks of the heart sounds from the homomorphic envelope
- Parameters:
signal (array) – Input filtered PCG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
- Returns:
peaks (array) – peak location indices.
envelope (array) – Homomorphic envelope (normalized).
- biosppy.signals.pcg.get_avg_heart_rate(envelope=None, sampling_rate=1000.0)[source]
Compute average heart rate from the signal’s homomorphic envelope.
Follows the approach described by Schmidt et al. [Schimdt10], with code adapted from David Springer [Springer16].
- Parameters:
envelope (array) – Signal’s homomorphic envelope
sampling_rate (int, float, optional) – Sampling frequency (Hz).
- Returns:
heart_rate (double) – Average heart rate (bpm).
systolic_time_interval (double) – Average systolic time interval (seconds).
Notes
Assumes normal human heart rate to be between 40 and 200 bpm.
Assumes normal human systole time interval to be between 0.2 seconds and half a heartbeat
References
[Schimdt10]S. E. Schmidt et al., “Segmentation of heart sound recordings by a duration-dependent hidden Markov model”, Physiol. Meas., 2010
[Springer16]D.Springer, “Heart sound segmentation code based on duration-dependant HMM”, 2016. Available at: https://github.com/davidspringer/Springer-Segmentation-Code
- biosppy.signals.pcg.homomorphic_filter(signal=None, sampling_rate=1000.0, f_LPF=8, order=2)[source]
Finds the homomorphic envelope of a signal.
Adapted to Python from original MATLAB code written by David Springer, 2016 (C), for comparison purposes in the paper [Springer15]. Available at: https://github.com/davidspringer/Springer-Segmentation-Code
Follows the approach described by Schmidt et al. [Schimdt10].
- Parameters:
signal (array) – Input filtered PCG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
f_LPF (int, float, optional) – Low pass cut-off frequency (Hz)
order (int, optional) – Order of Butterworth low pass filter.
- Returns:
envelope (array) – Homomorphic envelope (non-normalized).
References
[Springer15]D.Springer, “Logistic Regression-HSMM-based Heart Sound Segmentation”, IEEE Trans. Biomed. Eng., In Press, 2015.
[Schimdt10]S. E. Schmidt et al., “Segmentation of heart sound recordings by a duration-dependent hidden Markov model”, Physiol. Meas., 2010
- biosppy.signals.pcg.identify_heart_sounds(beats=None, sampling_rate=1000.0)[source]
Classify heart sound peaks as S1 or S2
- Parameters:
beats (array) – Peaks of heart sounds
sampling_rate (int, float, optional) – Sampling frequency (Hz).
- Returns:
classification (array) – Classification of heart sound peaks. 1 is S1, 2 is S2
- biosppy.signals.pcg.pcg(signal=None, sampling_rate=1000.0, units=None, path=None, show=True)[source]
- Parameters:
signal (array) – Raw PCG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered PCG signal.
peaks (array) – Peak location indices.
hs (array) – Classification of peaks as S1 or S2.
heart_rate (double) – Average heart rate (bpm).
systolic_time_interval (double) – Average systolic time interval (seconds).
heart_rate_ts (array) – Heart rate time axis reference (seconds).
inst_heart_rate (array) – Instantaneous heart rate (bpm).
biosppy.signals.ppg
This module provides methods to process Photoplethysmogram (PPG) signals.
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.ppg.find_onsets_elgendi2013(signal=None, sampling_rate=1000.0, peakwindow=0.111, beatwindow=0.667, beatoffset=0.02, mindelay=0.3)[source]
Determines onsets of PPG pulses.
- Parameters:
signal (array) – Input filtered PPG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
peakwindow (float) – Parameter W1 on referenced article Optimized at 0.111
beatwindow (float) – Parameter W2 on referenced article Optimized at 0.667
beatoffset (float) – Parameter beta on referenced article Optimized at 0.2
mindelay (float) – Minimum delay between peaks. Avoids false positives
- Returns:
onsets (array) – Indices of PPG pulse onsets.
params (dict) – Input parameters of the function
References
Elgendi M, Norton I, Brearley M, Abbott D, Schuurmans D (2013) Systolic Peak Detection in
Acceleration Photoplethysmograms Measured from Emergency Responders in Tropical Conditions. PLoS ONE 8(10): e76585. doi:10.1371/journal.pone.0076585.
Notes
Optimal ranges for signal filtering (from Elgendi et al. 2013): “Optimization of the beat detector’s spectral window for the lower frequency resulted in a value within 0.5– 1 Hz with the higher frequency within 7–15 Hz”
All the number references below between curly brackets {…} by the code refer to the line numbers of code in “Table 2 Algorithm IV: DETECTOR (PPG signal, F1, F2, W1, W2, b)” from Elgendi et al. 2013 for a better comparison of the algorithm
- biosppy.signals.ppg.find_onsets_kavsaoglu2016(signal=None, sampling_rate=1000.0, alpha=0.2, k=4, init_bpm=90, min_delay=0.6, max_BPM=150)[source]
Determines onsets of PPG pulses.
- Parameters:
signal (array) – Input filtered PPG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
alpha (float, optional) – Low-pass filter factor. Avoids abrupt changes of BPM.
k (int, float, optional) – Number of segments by pulse. Width of each segment = Period of pulse according to current BPM / k
init_bpm (int, float, optional) – Initial BPM. Higher value results in a smaller segment width.
min_delay (float) – Minimum delay between peaks as percentage of current BPM pulse period. Avoids false positives
max_bpm (int, float, optional) – Maximum BPM. Maximum value accepted as valid BPM.
- Returns:
onsets (array) – Indices of PPG pulse onsets.
window_marks (array) – Indices of segments window boundaries.
params (dict) – Input parameters of the function
References
Kavsaoğlu, Ahmet & Polat, Kemal & Bozkurt, Mehmet. (2016). An innovative peak detection algorithm for
photoplethysmography signals: An adaptive segmentation method. TURKISH JOURNAL OF ELECTRICAL ENGINEERING & COMPUTER SCIENCES. 24. 1782-1796. 10.3906/elk-1310-177.
Notes
This algorithm is an adaption of the one described on Kavsaoğlu et al. (2016). This version takes into account a minimum delay between peaks and builds upon the adaptive segmentation by using a low-pass filter for BPM changes. This way, even if the algorithm wrongly detects a peak, the BPM value will stay relatively constant so the next pulse can be correctly segmented.
- biosppy.signals.ppg.ppg(signal=None, sampling_rate=1000.0, units=None, show=True)[source]
Process a raw PPG signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw PPG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered PPG signal.
peaks (array) – Indices of PPG pulse peaks.
templates_ts (array) – Templates time axis reference (seconds).
templates (array) – Extracted heartbeat templates.
heart_rate_ts (array) – Heart rate time axis reference (seconds).
heart_rate (array) – Instantaneous heart rate (bpm).
- biosppy.signals.ppg.ppg_segmentation(signal=None, sampling_rate=1000.0, peaks=None, selection=False, peak_threshold=None)[source]
Segments a filtered PPG signal. Segmentation filtering is achieved by taking into account segments selected by peak height and pulse morphology.
- Parameters:
signal (array) – Filtered PPG signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
peaks (array) – List of PPG systolic peaks.
selection (bool, optional) – If True, performs selection with peak height and pulse morphology.
peak_threshold (int, float, optional) – If selection is True, selects peaks with height greater than defined threshold.
- Returns:
onsets (array) – Indices of PPG pulse onsets (i.e., start of beats) of the selected segments.
peaks (array) – List of PPG systolic peaks of the selected segments.
segments_loc (array) – Start and end indices for each selected pulse segment.
biosppy.signals.resp
This module provides methods to process Respiration (Resp) signals.
- copyright:
2015-2018 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- biosppy.signals.resp.resp(signal=None, sampling_rate=1000.0, units=None, path=None, show=True)[source]
Process a raw Respiration signal and extract relevant signal features using default parameters.
- Parameters:
signal (array) – Raw Respiration signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
units (str, optional) – The units of the input signal. If specified, the plot will have the y-axis labeled with the corresponding units.
path (str, optional) – If provided, the plot will be saved to the specified file.
show (bool, optional) – If True, show a summary plot.
- Returns:
ts (array) – Signal time axis reference (seconds).
filtered (array) – Filtered Respiration signal.
zeros (array) – Indices of Respiration zero crossings.
resp_rate_ts (array) – Respiration rate time axis reference (seconds).
resp_rate (array) – Instantaneous respiration rate (Hz).
biosppy.signals.tools
This module provides various signal analysis methods in the time and frequency domains.
- copyright:
2015-2023 by Instituto de Telecomunicacoes
- license:
BSD 3-clause, see LICENSE for more details.
- class biosppy.signals.tools.OnlineFilter(b=None, a=None)[source]
Bases:
objectOnline filtering.
- Parameters:
b (array) – Numerator coefficients.
a (array) – Denominator coefficients.
- biosppy.signals.tools.analytic_signal(signal=None, N=None)[source]
Compute analytic signal, using the Hilbert Transform.
- Parameters:
signal (array) – Input signal.
N (int, optional) – Number of Fourier components; default is len(signal).
- Returns:
amplitude (array) – Amplitude envelope of the analytic signal.
phase (array) – Instantaneous phase component of the analystic signal.
- biosppy.signals.tools.band_power(freqs=None, power=None, frequency=None, decibel=True)[source]
Compute the avearge power in a frequency band.
- Parameters:
freqs (array) – Array of frequencies (Hz) at which the power was computed.
power (array) – Input power spectrum.
frequency (list, array) – Pair of frequencies defining the band.
decibel (bool, optional) – If True, input power is in decibels.
- Returns:
avg_power (float) – The average power in the band.
- biosppy.signals.tools.detrend_smoothness_priors(signal, smoothing_factor=10)[source]
Detrending method based on smoothness priors applied to HRV signal analysis.
Follows the approach by Tarvainen et al. [Tarivainen2002].
- Parameters:
signal (array) – Signal to be detrended.
smoothing_factor (int, float, optional) – Smoothing parameter lambda. Default: 10.
- Returns:
detrended (array) – Detrended signal.
trend (array) – The trend component of the signal.
References
[Tarivainen2002]M. P. Tarvainen, P. O. Ranta-aho and P. A. Karjalainen, “An advanced detrending method with application to HRV analysis,” in IEEE Transactions on Biomedical Engineering, vol. 49, no. 2, pp. 172-175, Feb. 2002, doi: 10.1109/10.979357.
- biosppy.signals.tools.distance_profile(query=None, signal=None, metric='euclidean')[source]
Compute the distance profile of a query sequence against a signal.
Implements the algorithm described in [Mueen2014].
- Parameters:
query (array) – Input query signal sequence.
signal (array) – Input target time series signal.
metric (str, optional) – The distance metric to use; one of ‘euclidean’ or ‘pearson’; default is ‘euclidean’.
- Returns:
dist (array) – Distance of the query sequence to every sub-sequnce in the signal.
Notes
Computes distances on z-normalized data.
References
[Mueen2014]Abdullah Mueen, Hossein Hamooni, “Trilce Estrada: Time Series Join on Subsequence Correlation”, ICDM 2014: 450-459
- biosppy.signals.tools.filter_signal(signal=None, ftype='FIR', band='lowpass', order=None, frequency=None, sampling_rate=1000.0, **kwargs)[source]
Filter a signal according to the given parameters.
- Parameters:
signal (array) – Signal to filter.
ftype (str) –
- Filter type:
Finite Impulse Response filter (‘FIR’);
Butterworth filter (‘butter’);
Chebyshev filters (‘cheby1’, ‘cheby2’);
Elliptic filter (‘ellip’);
Bessel filter (‘bessel’).
Notch filter (‘notch’).
band (str) –
- Band type:
Low-pass filter (‘lowpass’);
High-pass filter (‘highpass’);
Band-pass filter (‘bandpass’);
Band-stop filter (‘bandstop’).
order (int) – Order of the filter.
frequency (int, float, list, array) –
- Cutoff frequencies; format depends on type of band:
’lowpass’ or ‘bandpass’: single frequency;
’bandpass’ or ‘bandstop’: pair of frequencies.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
**kwargs (dict, optional) –
Additional keyword arguments are passed to the underlying scipy.signal function. - Q : float
Quality factor (only for ‘notch’ filter). Default: 30.
- Returns:
signal (array) – Filtered signal.
sampling_rate (float) – Sampling frequency (Hz).
params (dict) – Filter parameters.
Notes
Uses a forward-backward filter implementation. Therefore, the combined filter has linear phase.
- biosppy.signals.tools.find_extrema(signal=None, mode='both')[source]
Locate local extrema points in a signal.
Based on Fermat’s Theorem [Ferm].
- Parameters:
signal (array) – Input signal.
mode (str, optional) – Whether to find maxima (‘max’), minima (‘min’), or both (‘both’).
- Returns:
extrema (array) – Indices of the extrema points.
values (array) – Signal values at the extrema points.
References
[Ferm]Wikipedia, “Fermat’s theorem (stationary points)”, https://en.wikipedia.org/wiki/Fermat%27s_theorem_(stationary_points)
- biosppy.signals.tools.find_intersection(x1=None, y1=None, x2=None, y2=None, alpha=1.5, xtol=1e-06, ytol=1e-06)[source]
Find the intersection points between two lines using piecewise polynomial interpolation.
- Parameters:
x1 (array) – Array of x-coordinates of the first line.
y1 (array) – Array of y-coordinates of the first line.
x2 (array) – Array of x-coordinates of the second line.
y2 (array) – Array of y-coordinates of the second line.
alpha (float, optional) – Resolution factor for the x-axis; fraction of total number of x-coordinates.
xtol (float, optional) – Tolerance for the x-axis.
ytol (float, optional) – Tolerance for the y-axis.
- Returns:
roots (array) – Array of x-coordinates of found intersection points.
values (array) – Array of y-coordinates of found intersection points.
Notes
If no intersection is found, returns the closest point.
- biosppy.signals.tools.finite_difference(signal=None, weights=None)[source]
Apply the Finite Difference method to compute derivatives.
- Parameters:
signal (array) – Signal to differentiate.
weights (list, array) – Finite difference weight coefficients.
- Returns:
index (array) – Indices from signal for which the derivative was computed.
derivative (array) – Computed derivative.
Notes
The method assumes central differences weights.
The method accounts for the delay introduced by the algorithm.
- Raises:
ValueError – If the number of weights is not odd.
- biosppy.signals.tools.get_filter(ftype='FIR', band='lowpass', order=None, frequency=None, sampling_rate=1000.0, **kwargs)[source]
Compute digital (FIR or IIR) filter coefficients with the given parameters.
- Parameters:
ftype (str) –
- Filter type:
Finite Impulse Response filter (‘FIR’);
Butterworth filter (‘butter’);
Chebyshev filters (‘cheby1’, ‘cheby2’);
Elliptic filter (‘ellip’);
Bessel filter (‘bessel’).
Notch filter (‘notch’).
band (str) –
- Band type:
Low-pass filter (‘lowpass’);
High-pass filter (‘highpass’);
Band-pass filter (‘bandpass’);
Band-stop filter (‘bandstop’).
order (int) – Order of the filter.
frequency (int, float, list, array) –
- Cutoff frequencies; format depends on type of band:
’lowpass’ or ‘highpass’: single frequency;
’bandpass’ or ‘bandstop’: pair of frequencies.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
**kwargs (dict, optional) –
Additional keyword arguments are passed to the underlying scipy.signal function. - Q : float
Quality factor (only for ‘notch’ filter). Default: 30.
- Returns:
b (array) – Numerator coefficients.
a (array) – Denominator coefficients.
See Also – scipy.signal
- biosppy.signals.tools.get_heart_rate(beats=None, sampling_rate=1000.0, smooth=False, size=3)[source]
Compute instantaneous heart rate from an array of beat indices.
- Parameters:
beats (array) – Beat location indices.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
smooth (bool, optional) – If True, perform smoothing on the resulting heart rate.
size (int, optional) – Size of smoothing window; ignored if smooth is False.
- Returns:
index (array) – Heart rate location indices.
heart_rate (array) – Instantaneous heart rate (bpm).
Notes
Assumes normal human heart rate to be between 40 and 200 bpm.
- biosppy.signals.tools.mean_waves(data=None, size=None, step=None)[source]
Extract mean samples from a data set.
- Parameters:
data (array) – An m by n array of m data samples in an n-dimensional space.
size (int) – Number of samples to use for each mean sample.
step (int, optional) – Number of samples to jump, controlling overlap; default is equal to size (no overlap).
- Returns:
waves (array) – An k by n array of mean samples.
Notes
Discards trailing samples if they are not enough to satify the size parameter.
- Raises:
ValueError – If step is an invalid value.
ValueError – If there are not enough samples for the given size.
- biosppy.signals.tools.median_waves(data=None, size=None, step=None)[source]
Extract median samples from a data set.
- Parameters:
data (array) – An m by n array of m data samples in an n-dimensional space.
size (int) – Number of samples to use for each median sample.
step (int, optional) – Number of samples to jump, controlling overlap; default is equal to size (no overlap).
- Returns:
waves (array) – An k by n array of median samples.
Notes
Discards trailing samples if they are not enough to satify the size parameter.
- Raises:
ValueError – If step is an invalid value.
ValueError – If there are not enough samples for the given size.
- biosppy.signals.tools.normalize(signal=None, ddof=1)[source]
Normalize a signal to zero mean and unitary standard deviation.
- Parameters:
signal (array) – Input signal.
ddof (int, optional) – Delta degrees of freedom for standard deviation computation; the divisor is N - ddof, where N is the number of elements; default is one.
- Returns:
signal (array) – Normalized signal.
- biosppy.signals.tools.pearson_correlation(x=None, y=None)[source]
Compute the Pearson Correlation Coefficient bertween two signals.
The coefficient is given by:
![r_{xy} = \frac{E[(X - \mu_X) (Y - \mu_Y)]}{\sigma_X \sigma_Y}](_images/math/25984ac0b99af0b57d7d197c63928096548adcb5.png)
- Parameters:
x (array) – First input signal.
y (array) – Second input signal.
- Returns:
rxy (float) – Pearson correlation coefficient, ranging between -1 and +1.
- Raises:
ValueError – If the input signals do not have the same length.
- biosppy.signals.tools.phase_locking(signal1=None, signal2=None, N=None)[source]
Compute the Phase-Locking Factor (PLF) between two signals.
- Parameters:
signal1 (array) – First input signal.
signal2 (array) – Second input signal.
N (int, optional) – Number of Fourier components.
- Returns:
plf (float) – The PLF between the two signals.
- biosppy.signals.tools.power_spectrum(signal=None, sampling_rate=1000.0, pad=None, pow2=False, decibel=True)[source]
Compute the power spectrum of a signal (one-sided).
- Parameters:
signal (array) – Input signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
pad (int, optional) – Padding for the Fourier Transform (number of zeros added); defaults to no padding..
pow2 (bool, optional) – If True, rounds the number of points N = len(signal) + pad to the nearest power of 2 greater than N.
decibel (bool, optional) – If True, returns the power in decibels.
- Returns:
freqs (array) – Array of frequencies (Hz) at which the power was computed.
power (array) – Power spectrum.
- biosppy.signals.tools.rms_error(x=None, y=None)[source]
Compute the Root-Mean-Square Error between two signals.
The error is given by:
![rmse = \sqrt{E[(X - Y)^2]}](_images/math/54caf68afcbb74e741a359c4a7afb94b23e61956.png)
- Parameters:
x (array) – First input signal.
y (array) – Second input signal.
- Returns:
rmse (float) – Root-mean-square error.
- Raises:
ValueError – If the input signals do not have the same length.
- biosppy.signals.tools.signal_cross_join(signal1=None, signal2=None, size=None, index=None, limit=None)[source]
Compute the matrix profile for a similarity join of two time series.
Computes the nearest sub-sequence in signal2 for each sub-sequence in signal1. Implements the algorithm described in [Yeh2016_c].
- Parameters:
signal1 (array) – Fisrt input time series signal.
signal2 (array) – Second input time series signal.
size (int) – Size of the query sub-sequences.
index (list, array, optional) – Starting indices for query sub-sequences; the default is to search all sub-sequences.
limit (int, optional) – Upper limit for the number of query sub-sequences; the default is to search all sub-sequences.
- Returns:
matrix_index (array) – Matric profile index.
matrix_profile (array) – Computed matrix profile (distances).
Notes
Computes euclidean distances on z-normalized data.
References
[Yeh2016_c]Chin-Chia Michael Yeh, Yan Zhu, Liudmila Ulanova, Nurjahan Begum, Yifei Ding, Hoang Anh Dau, Diego Furtado Silva, Abdullah Mueen, Eamonn Keogh, “Matrix Profile I: All Pairs Similarity Joins for Time Series: A Unifying View that Includes Motifs, Discords and Shapelets”, IEEE ICDM 2016
- biosppy.signals.tools.signal_self_join(signal=None, size=None, index=None, limit=None)[source]
Compute the matrix profile for a self-similarity join of a time series.
Implements the algorithm described in [Yeh2016_b].
- Parameters:
signal (array) – Input target time series signal.
size (int) – Size of the query sub-sequences.
index (list, array, optional) – Starting indices for query sub-sequences; the default is to search all sub-sequences.
limit (int, optional) – Upper limit for the number of query sub-sequences; the default is to search all sub-sequences.
- Returns:
matrix_index (array) – Matric profile index.
matrix_profile (array) – Computed matrix profile (distances).
Notes
Computes euclidean distances on z-normalized data.
References
[Yeh2016_b]Chin-Chia Michael Yeh, Yan Zhu, Liudmila Ulanova, Nurjahan Begum, Yifei Ding, Hoang Anh Dau, Diego Furtado Silva, Abdullah Mueen, Eamonn Keogh, “Matrix Profile I: All Pairs Similarity Joins for Time Series: A Unifying View that Includes Motifs, Discords and Shapelets”, IEEE ICDM 2016
- biosppy.signals.tools.signal_stats(signal=None)[source]
Compute various metrics describing the signal.
- Parameters:
signal (array) – Input signal.
- Returns:
mean (float) – Mean of the signal.
median (float) – Median of the signal.
min (float) – Minimum signal value.
max (float) – Maximum signal value.
max_amp (float) – Maximum absolute signal amplitude, in relation to the mean.
range (float) – Signal range (max - min).
q1 (float) – First quartile of the signal.
q3 (float) – Third quartile of the signal.
var (float) – Signal variance (unbiased).
std_dev (float) – Standard signal deviation (unbiased).
abs_dev (float) – Mean absolute signal deviation around the median.
rms (float) – Root-mean-square of the signal.
kurtosis (float) – Signal kurtosis (unbiased).
skew (float) – Signal skewness (unbiased).
- biosppy.signals.tools.smoother(signal=None, kernel='boxzen', size=10, mirror=True, **kwargs)[source]
Smooth a signal using an N-point moving average [MAvg] filter.
This implementation uses the convolution of a filter kernel with the input signal to compute the smoothed signal [Smit97].
Availabel kernels: median, boxzen, boxcar, triang, blackman, hamming, hann, bartlett, flattop, parzen, bohman, blackmanharris, nuttall, barthann, kaiser (needs beta), gaussian (needs std), general_gaussian (needs power, width), slepian (needs width), chebwin (needs attenuation).
- Parameters:
signal (array) – Signal to smooth.
kernel (str, array, optional) – Type of kernel to use; if array, use directly as the kernel.
size (int, optional) – Size of the kernel; ignored if kernel is an array.
mirror (bool, optional) – If True, signal edges are extended to avoid boundary effects.
**kwargs (dict, optional) – Additional keyword arguments are passed to the underlying scipy.signal.windows function.
- Returns:
signal (array) – Smoothed signal.
params (dict) – Smoother parameters.
Notes
When the kernel is ‘median’, mirror is ignored.
References
[MAvg]Wikipedia, “Moving Average”, http://en.wikipedia.org/wiki/Moving_average
[Smit97]S. W. Smith, “Moving Average Filters - Implementation by Convolution”, http://www.dspguide.com/ch15/1.htm, 1997
- biosppy.signals.tools.synchronize(x=None, y=None, detrend=True)[source]
Align two signals based on cross-correlation.
- Parameters:
x (array) – First input signal.
y (array) – Second input signal.
detrend (bool, optional) – If True, remove signal means before computation.
- Returns:
delay (int) – Delay (number of samples) of ‘x’ in relation to ‘y’; if ‘delay’ < 0 , ‘x’ is ahead in relation to ‘y’; if ‘delay’ > 0 , ‘x’ is delayed in relation to ‘y’.
corr (float) – Value of maximum correlation.
synch_x (array) – Biggest possible portion of ‘x’ in synchronization.
synch_y (array) – Biggest possible portion of ‘y’ in synchronization.
- biosppy.signals.tools.welch_spectrum(signal=None, sampling_rate=1000.0, size=None, overlap=None, window='hanning', window_kwargs=None, pad=None, decibel=True)[source]
Compute the power spectrum of a signal using Welch’s method (one-sided).
- Parameters:
signal (array) – Input signal.
sampling_rate (int, float, optional) – Sampling frequency (Hz).
size (int, optional) – Number of points in each Welch segment; defaults to the equivalent of 1 second; ignored when ‘window’ is an array.
overlap (int, optional) – Number of points to overlap between segments; defaults to size / 2.
window (str, array, optional) – Type of window to use.
window_kwargs (dict, optional) – Additional keyword arguments to pass on window creation; ignored if ‘window’ is an array.
pad (int, optional) – Padding for the Fourier Transform (number of zeros added); defaults to no padding.
decibel (bool, optional) – If True, returns the power in decibels.
- Returns:
freqs (array) – Array of frequencies (Hz) at which the power was computed.
power (array) – Power spectrum.
Notes
Detrends each Welch segment by removing the mean.
- biosppy.signals.tools.windower(signal=None, size=None, step=None, fcn=None, fcn_kwargs=None, kernel='boxcar', kernel_kwargs=None)[source]
Apply a function to a signal in sequential windows, with optional overlap.
Availabel window kernels: boxcar, triang, blackman, hamming, hann, bartlett, flattop, parzen, bohman, blackmanharris, nuttall, barthann, kaiser (needs beta), gaussian (needs std), general_gaussian (needs power, width), slepian (needs width), chebwin (needs attenuation).
- Parameters:
signal (array) – Input signal.
size (int) – Size of the signal window.
step (int, optional) – Size of window shift; if None, there is no overlap.
fcn (callable) – Function to apply to each window.
fcn_kwargs (dict, optional) – Additional keyword arguments to pass to ‘fcn’.
kernel (str, array, optional) – Type of kernel to use; if array, use directly as the kernel.
kernel_kwargs (dict, optional) – Additional keyword arguments to pass on window creation; ignored if ‘kernel’ is an array.
- Returns:
index (array) – Indices characterizing window locations (start of the window).
values (array) – Concatenated output of calling ‘fcn’ on each window.
- biosppy.signals.tools.zero_cross(signal=None, detrend=False)[source]
Locate the indices where the signal crosses zero.
- Parameters:
signal (array) – Input signal.
detrend (bool, optional) – If True, remove signal mean before computation.
- Returns:
zeros (array) – Indices of zero crossings.
Notes
When the signal crosses zero between samples, the first index is returned.