biosppy.signals¶
This sub-package provides methods to process common physiological signals (biosignals).
Modules¶
biosppy.signals.bvp¶
This module provides methods to process Blood Volume Pulse (BVP) signals.
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
biosppy.signals.bvp.bvp(signal=None, sampling_rate=1000.0, show=True)¶ Process a raw BVP signal and extract relevant signal features using default parameters.
Parameters: - signal (array) – Raw BVP signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- show (bool, optional) – If True, show a summary plot.
Returns: - ts (array) – Signal time axis reference (seconds).
- filtered (array) – Filtered BVP signal.
- onsets (array) – Indices of BVP pulse onsets.
- heart_rate_ts (array) – Heart rate time axis reference (seconds).
- heart_rate (array) – Instantaneous heart rate (bpm).
-
biosppy.signals.bvp.find_onsets(signal=None, sampling_rate=1000.0, sm_size=None, size=None, alpha=2.0, wrange=None, d1_th=0, d2_th=None)¶ Determine onsets of BVP pulses.
Skips corrupted signal parts. Based on the approach by Zong et al. [Zong03].
Parameters: - signal (array) – Input filtered BVP signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- sm_size (int, optional) – Size of smoother kernel (seconds). Defaults to 0.25
- size (int, optional) – Window to search for maxima (seconds). Defaults to 5
- alpha (float, optional) – Normalization parameter. Defaults to 2.0
- wrange (int, optional) – The window in which to search for a peak (seconds). Defaults to 0.1
- d1_th (int, optional) – Smallest allowed difference between maxima and minima. Defaults to 0
- d2_th (int, optional) – Smallest allowed time between maxima and minima (seconds), Defaults to 0.15
Returns: onsets (array) – Indices of BVP pulse onsets.
References
[Zong03] W Zong, T Heldt, GB Moody and RG Mark, “An Open-source Algorithm to Detect Onset of Arterial Blood Pressure Pulses”, IEEE Comp. in Cardiology, vol. 30, pp. 259-262, 2003
biosppy.signals.ecg¶
This module provides methods to process Electrocardiographic (ECG) signals. Implemented code assumes a single-channel Lead I like ECG signal.
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
biosppy.signals.ecg.christov_segmenter(signal=None, sampling_rate=1000.0)¶ ECG R-peak segmentation algorithm.
Follows the approach by Christov [Chri04].
Parameters: - signal (array) – Input filtered ECG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
Returns: rpeaks (array) – R-peak location indices.
References
[Chri04] Ivaylo I. Christov, “Real time electrocardiogram QRS detection using combined adaptive threshold”, BioMedical Engineering OnLine 2004, vol. 3:28, 2004
-
biosppy.signals.ecg.compare_segmentation(reference=None, test=None, sampling_rate=1000.0, offset=0, minRR=None, tol=0.05)¶ Compare the segmentation performance of a list of R-peak positions against a reference list.
Parameters: - reference (array) – Reference R-peak location indices.
- test (array) – Test R-peak location indices.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- offset (int, optional) – Constant a priori offset (number of samples) between reference and test R-peak locations.
- minRR (float, optional) – Minimum admissible RR interval (seconds).
- tol (float, optional) – Tolerance between corresponding reference and test R-peak locations (seconds).
Returns: - TP (int) – Number of true positive R-peaks.
- FP (int) – Number of false positive R-peaks.
- performance (float) – Test performance; TP / len(reference).
- acc (float) – Accuracy rate; TP / (TP + FP).
- err (float) – Error rate; FP / (TP + FP).
- match (list) – Indices of the elements of ‘test’ that match to an R-peak from ‘reference’.
- deviation (array) – Absolute errors of the matched R-peaks (seconds).
- mean_deviation (float) – Mean error (seconds).
- std_deviation (float) – Standard deviation of error (seconds).
- mean_ref_ibi (float) – Mean of the reference interbeat intervals (seconds).
- std_ref_ibi (float) – Standard deviation of the reference interbeat intervals (seconds).
- mean_test_ibi (float) – Mean of the test interbeat intervals (seconds).
- std_test_ibi (float) – Standard deviation of the test interbeat intervals (seconds).
-
biosppy.signals.ecg.correct_rpeaks(signal=None, rpeaks=None, sampling_rate=1000.0, tol=0.05)¶ Correct R-peak locations to the maximum within a tolerance.
Parameters: - signal (array) – ECG signal.
- rpeaks (array) – R-peak location indices.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- tol (int, float, optional) – Correction tolerance (seconds).
Returns: rpeaks (array) – Cerrected R-peak location indices.
Notes
- The tolerance is defined as the time interval
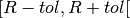 .
.
-
biosppy.signals.ecg.ecg(signal=None, sampling_rate=1000.0, show=True)¶ Process a raw ECG signal and extract relevant signal features using default parameters.
Parameters: - signal (array) – Raw ECG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- show (bool, optional) – If True, show a summary plot.
Returns: - ts (array) – Signal time axis reference (seconds).
- filtered (array) – Filtered ECG signal.
- rpeaks (array) – R-peak location indices.
- templates_ts (array) – Templates time axis reference (seconds).
- templates (array) – Extracted heartbeat templates.
- heart_rate_ts (array) – Heart rate time axis reference (seconds).
- heart_rate (array) – Instantaneous heart rate (bpm).
-
biosppy.signals.ecg.engzee_segmenter(signal=None, sampling_rate=1000.0, threshold=0.48)¶ ECG R-peak segmentation algorithm.
Follows the approach by Engelse and Zeelenberg [EnZe79] with the modifications by Lourenco et al. [LSLL12].
Parameters: - signal (array) – Input filtered ECG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- threshold (float, optional) – Detection threshold.
Returns: rpeaks (array) – R-peak location indices.
References
[EnZe79] W. Engelse and C. Zeelenberg, “A single scan algorithm for QRS detection and feature extraction”, IEEE Comp. in Cardiology, vol. 6, pp. 37-42, 1979 [LSLL12] A. Lourenco, H. Silva, P. Leite, R. Lourenco and A. Fred, “Real Time Electrocardiogram Segmentation for Finger Based ECG Biometrics”, BIOSIGNALS 2012, pp. 49-54, 2012
-
biosppy.signals.ecg.extract_heartbeats(signal=None, rpeaks=None, sampling_rate=1000.0, before=0.2, after=0.4)¶ Extract heartbeat templates from an ECG signal, given a list of R-peak locations.
Parameters: - signal (array) – Input ECG signal.
- rpeaks (array) – R-peak location indices.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- before (float, optional) – Window size to include before the R peak (seconds).
- after (int, optional) – Window size to include after the R peak (seconds).
Returns: - templates (array) – Extracted heartbeat templates.
- rpeaks (array) – Corresponding R-peak location indices of the extracted heartbeat templates.
-
biosppy.signals.ecg.gamboa_segmenter(signal=None, sampling_rate=1000.0, tol=0.002)¶ ECG R-peak segmentation algorithm.
Follows the approach by Gamboa.
Parameters: - signal (array) – Input filtered ECG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- tol (float, optional) – Tolerance parameter.
Returns: rpeaks (array) – R-peak location indices.
-
biosppy.signals.ecg.hamilton_segmenter(signal=None, sampling_rate=1000.0)¶ ECG R-peak segmentation algorithm.
Follows the approach by Hamilton [Hami02].
Parameters: - signal (array) – Input filtered ECG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
Returns: rpeaks (array) – R-peak location indices.
References
[Hami02] P.S. Hamilton, “Open Source ECG Analysis Software Documentation”, E.P.Limited, 2002
-
biosppy.signals.ecg.ssf_segmenter(signal=None, sampling_rate=1000.0, threshold=20, before=0.03, after=0.01)¶ ECG R-peak segmentation based on the Slope Sum Function (SSF).
Parameters: - signal (array) – Input filtered ECG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- threshold (float, optional) – SSF threshold.
- before (float, optional) – Search window size before R-peak candidate (seconds).
- after (float, optional) – Search window size after R-peak candidate (seconds).
Returns: rpeaks (array) – R-peak location indices.
biosppy.signals.eda¶
This module provides methods to process Electrodermal Activity (EDA) signals, also known as Galvanic Skin Response (GSR).
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
biosppy.signals.eda.basic_scr(signal=None, sampling_rate=1000.0)¶ Basic method to extract Skin Conductivity Responses (SCR) from an EDA signal.
Follows the approach in [Gamb08].
Parameters: - signal (array) – Input filterd EDA signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
Returns: - onsets (array) – Indices of the SCR onsets.
- peaks (array) – Indices of the SRC peaks.
- amplitudes (array) – SCR pulse amplitudes.
References
[Gamb08] Hugo Gamboa, “Multi-modal Behavioral Biometrics Based on HCI and Electrophysiology”, PhD thesis, Instituto Superior T{‘e}cnico, 2008
-
biosppy.signals.eda.eda(signal=None, sampling_rate=1000.0, show=True, min_amplitude=0.1)¶ Process a raw EDA signal and extract relevant signal features using default parameters.
Parameters: - signal (array) – Raw EDA signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- show (bool, optional) – If True, show a summary plot.
- min_amplitude (float, optional) – Minimum treshold by which to exclude SCRs.
Returns: - ts (array) – Signal time axis reference (seconds).
- filtered (array) – Filtered EDA signal.
- onsets (array) – Indices of SCR pulse onsets.
- peaks (array) – Indices of the SCR peaks.
- amplitudes (array) – SCR pulse amplitudes.
-
biosppy.signals.eda.kbk_scr(signal=None, sampling_rate=1000.0, min_amplitude=0.1)¶ KBK method to extract Skin Conductivity Responses (SCR) from an EDA signal.
Follows the approach by Kim et al. [KiBK04].
Parameters: - signal (array) – Input filterd EDA signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- min_amplitude (float, optional) – Minimum treshold by which to exclude SCRs.
Returns: - onsets (array) – Indices of the SCR onsets.
- peaks (array) – Indices of the SRC peaks.
- amplitudes (array) – SCR pulse amplitudes.
References
[KiBK04] K.H. Kim, S.W. Bang, and S.R. Kim, “Emotion recognition system using short-term monitoring of physiological signals”, Med. Biol. Eng. Comput., vol. 42, pp. 419-427, 2004
biosppy.signals.eeg¶
This module provides methods to process Electroencephalographic (EEG) signals.
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
biosppy.signals.eeg.car_reference(signal=None)¶ Change signal reference to the Common Average Reference (CAR).
Parameters: signal (array) – Input EEG signal matrix; each column is one EEG channel. Returns: signal (array) – Re-referenced EEG signal matrix; each column is one EEG channel.
-
biosppy.signals.eeg.eeg(signal=None, sampling_rate=1000.0, labels=None, show=True)¶ Process raw EEG signals and extract relevant signal features using default parameters.
Parameters: - signal (array) – Raw EEG signal matrix; each column is one EEG channel.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- labels (list, optional) – Channel labels.
- show (bool, optional) – If True, show a summary plot.
Returns: - ts (array) – Signal time axis reference (seconds).
- filtered (array) – Filtered BVP signal.
- features_ts (array) – Features time axis reference (seconds).
- theta (array) – Average power in the 4 to 8 Hz frequency band; each column is one EEG channel.
- alpha_low (array) – Average power in the 8 to 10 Hz frequency band; each column is one EEG channel.
- alpha_high (array) – Average power in the 10 to 13 Hz frequency band; each column is one EEG channel.
- beta (array) – Average power in the 13 to 25 Hz frequency band; each column is one EEG channel.
- gamma (array) – Average power in the 25 to 40 Hz frequency band; each column is one EEG channel.
- plf_pairs (list) – PLF pair indices.
- plf (array) – PLF matrix; each column is a channel pair.
-
biosppy.signals.eeg.get_plf_features(signal=None, sampling_rate=1000.0, size=0.25, overlap=0.5)¶ Extract Phase-Locking Factor (PLF) features from EEG signals between all channel pairs.
Parameters: - signal (array) – Filtered EEG signal matrix; each column is one EEG channel.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (float, optional) – Window size (seconds).
- overlap (float, optional) – Window overlap (0 to 1).
Returns: - ts (array) – Features time axis reference (seconds).
- plf_pairs (list) – PLF pair indices.
- plf (array) – PLF matrix; each column is a channel pair.
-
biosppy.signals.eeg.get_power_features(signal=None, sampling_rate=1000.0, size=0.25, overlap=0.5)¶ Extract band power features from EEG signals.
Computes the average signal power, with overlapping windows, in typical EEG frequency bands: * Theta: from 4 to 8 Hz, * Lower Alpha: from 8 to 10 Hz, * Higher Alpha: from 10 to 13 Hz, * Beta: from 13 to 25 Hz, * Gamma: from 25 to 40 Hz.
Parameters: - array (signal) – Filtered EEG signal matrix; each column is one EEG channel.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (float, optional) – Window size (seconds).
- overlap (float, optional) – Window overlap (0 to 1).
Returns: - ts (array) – Features time axis reference (seconds).
- theta (array) – Average power in the 4 to 8 Hz frequency band; each column is one EEG channel.
- alpha_low (array) – Average power in the 8 to 10 Hz frequency band; each column is one EEG channel.
- alpha_high (array) – Average power in the 10 to 13 Hz frequency band; each column is one EEG channel.
- beta (array) – Average power in the 13 to 25 Hz frequency band; each column is one EEG channel.
- gamma (array) – Average power in the 25 to 40 Hz frequency band; each column is one EEG channel.
biosppy.signals.emg¶
This module provides methods to process Electromyographic (EMG) signals.
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
biosppy.signals.emg.abbink_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, alarm_size=None, threshold=None, transition_threshold=None)¶ Determine onsets of EMG pulses.
Follows the approach by Abbink et al.. [Abb98].
Parameters: - signal (array) – Input filtered EMG signal.
- rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (int) – Detection window size (seconds).
- alarm_size (int) – Number of amplitudes searched in the calculation of the transition index.
- threshold (int, float) – Detection threshold.
- transition_threshold (int, float) – Threshold used in the calculation of the transition index.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[Abb98] Abbink JH, van der Bilt A, van der Glas HW, “Detection of onset and termination of muscle activity in surface electromyograms”, Journal of Oral Rehabilitation, vol. 25, pp. 365–369, 1998
-
biosppy.signals.emg.bonato_onset_detector(signal=None, rest=None, sampling_rate=1000.0, threshold=None, active_state_duration=None, samples_above_fail=None, fail_size=None)¶ Determine onsets of EMG pulses.
Follows the approach by Bonato et al. [Bo98].
Parameters: - signal (array) – Input filtered EMG signal.
- rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- threshold (int, float) – Detection threshold.
- active_state_duration (int) – Minimum duration of the active state.
- samples_above_fail (int) – Number of samples above the threshold level in a group of successive samples.
- fail_size (int) – Number of successive samples.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[Bo98] Bonato P, D’Alessio T, Knaflitz M, “A statistical method for the measurement of muscle activation intervals from surface myoelectric signal during gait”, IEEE Transactions on Biomedical Engineering, vol. 45:3, pp. 287–299, 1998
-
biosppy.signals.emg.emg(signal=None, sampling_rate=1000.0, show=True)¶ Process a raw EMG signal and extract relevant signal features using default parameters.
Parameters: - signal (array) – Raw EMG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- show (bool, optional) – If True, show a summary plot.
Returns: - ts (array) – Signal time axis reference (seconds).
- filtered (array) – Filtered EMG signal.
- onsets (array) – Indices of EMG pulse onsets.
-
biosppy.signals.emg.find_onsets(signal=None, sampling_rate=1000.0, size=0.05, threshold=None)¶ Determine onsets of EMG pulses.
Skips corrupted signal parts.
Parameters: - signal (array) – Input filtered EMG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (float, optional) – Detection window size (seconds).
- threshold (float, optional) – Detection threshold.
Returns: onsets (array) – Indices of EMG pulse onsets.
-
biosppy.signals.emg.hodges_bui_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, threshold=None)¶ Determine onsets of EMG pulses.
Follows the approach by Hodges and Bui [HoBu96].
Parameters: - signal (array) – Input filtered EMG signal.
- rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (int) – Detection window size (seconds).
- threshold (int, float) – Detection threshold.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[HoBu96] Hodges PW, Bui BH, “A comparison of computer-based methods for the determination of onset of muscle contraction using electromyography”, Electroencephalography and Clinical Neurophysiology - Electromyography and Motor Control, vol. 101:6, pp. 511-519, 1996
-
biosppy.signals.emg.lidierth_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, threshold=None, active_state_duration=None, fail_size=None)¶ Determine onsets of EMG pulses.
Follows the approach by Lidierth. [Li86].
Parameters: - signal (array) – Input filtered EMG signal.
- rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (int) – Detection window size (seconds).
- threshold (int, float) – Detection threshold.
- active_state_duration (int) – Minimum duration of the active state.
- fail_size (int) – Number of successive samples.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[Li86] Lidierth M, “A computer based method for automated measurement of the periods of muscular activity from an EMG and its application to locomotor EMGs”, ElectroencephClin Neurophysiol, vol. 64:4, pp. 378–380, 1986
-
biosppy.signals.emg.londral_onset_detector(signal=None, rest=None, sampling_rate=1000.0, size=None, threshold=None, active_state_duration=None)¶ Determine onsets of EMG pulses.
Follows the approach by Londral et al. [Lon13].
Parameters: - signal (array) – Input filtered EMG signal.
- rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (int) – Detection window size (seconds).
- threshold (int, float) – Scale factor for calculating the detection threshold.
- active_state_duration (int) – Minimum duration of the active state.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[Lon13] Londral A, Silva H, Nunes N, Carvalho M, Azevedo L, “A wireless user-computer interface to explore various sources of biosignals and visual biofeedback for severe motor impairment”, Journal of Accessibility and Design for All, vol. 3:2, pp. 118–134, 2013
-
biosppy.signals.emg.silva_onset_detector(signal=None, sampling_rate=1000.0, size=None, threshold_size=None, threshold=None)¶ Determine onsets of EMG pulses.
Follows the approach by Silva et al. [Sil12].
Parameters: - signal (array) – Input filtered EMG signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (int) – Detection window size (seconds).
- threshold_size (int) – Window size for calculation of the adaptive threshold; must be bigger than the detection window size.
- threshold (int, float) – Fixed threshold for the double criteria.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[Sil12] Silva H, Scherer R, Sousa J, Londral A , “Towards improving the usability of electromyographic interfacess”, Journal of Oral Rehabilitation, pp. 1–2, 2012
-
biosppy.signals.emg.solnik_onset_detector(signal=None, rest=None, sampling_rate=1000.0, threshold=None, active_state_duration=None)¶ Determine onsets of EMG pulses.
Follows the approach by Solnik et al. [Sol10].
Parameters: - signal (array) – Input filtered EMG signal.
- rest (array, list, dict) – One of the following 3 options: * N-dimensional array with filtered samples corresponding to a rest period; * 2D array or list with the beginning and end indices of a segment of the signal corresponding to a rest period; * Dictionary with {‘mean’: mean value, ‘std_dev’: standard variation}.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- threshold (int, float) – Scale factor for calculating the detection threshold.
- active_state_duration (int) – Minimum duration of the active state.
Returns: - onsets (array) – Indices of EMG pulse onsets.
- processed (array) – Processed EMG signal.
References
[Sol10] Solnik S, Rider P, Steinweg K, DeVita P, Hortobágyi T, “Teager-Kaiser energy operator signal conditioning improves EMG onset detection”, European Journal of Applied Physiology, vol 110:3, pp. 489-498, 2010
biosppy.signals.resp¶
This module provides methods to process Respiration (Resp) signals.
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
biosppy.signals.resp.resp(signal=None, sampling_rate=1000.0, show=True)¶ Process a raw Respiration signal and extract relevant signal features using default parameters.
Parameters: - signal (array) – Raw Respiration signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- show (bool, optional) – If True, show a summary plot.
Returns: - ts (array) – Signal time axis reference (seconds).
- filtered (array) – Filtered Respiration signal.
- zeros (array) – Indices of Respiration zero crossings.
- resp_rate_ts (array) – Respiration rate time axis reference (seconds).
- resp_rate (array) – Instantaneous respiration rate (Hz).
biosppy.signals.tools¶
This module provides various signal analysis methods in the time and frequency domains.
| copyright: |
|
|---|---|
| license: | BSD 3-clause, see LICENSE for more details. |
-
class
biosppy.signals.tools.OnlineFilter(b=None, a=None)¶ Bases:
objectOnline filtering.
Parameters: - b (array) – Numerator coefficients.
- a (array) – Denominator coefficients.
-
filter(signal=None)¶ Filter a signal segment.
Parameters: signal (array) – Signal segment to filter. Returns: filtered (array) – Filtered signal segment.
-
reset()¶ Reset the filter state.
-
biosppy.signals.tools.analytic_signal(signal=None, N=None)¶ Compute analytic signal, using the Hilbert Transform.
Parameters: - signal (array) – Input signal.
- N (int, optional) – Number of Fourier components; default is len(signal).
Returns: - amplitude (array) – Amplitude envelope of the analytic signal.
- phase (array) – Instantaneous phase component of the analystic signal.
-
biosppy.signals.tools.band_power(freqs=None, power=None, frequency=None, decibel=True)¶ Compute the avearge power in a frequency band.
Parameters: - freqs (array) – Array of frequencies (Hz) at which the power was computed.
- power (array) – Input power spectrum.
- frequency (list, array) – Pair of frequencies defining the band.
- decibel (bool, optional) – If True, input power is in decibels.
Returns: avg_power (float) – The average power in the band.
-
biosppy.signals.tools.distance_profile(query=None, signal=None, metric='euclidean')¶ Compute the distance profile of a query sequence against a signal.
Implements the algorithm described in [Mueen2014].
Parameters: - query (array) – Input query signal sequence.
- signal (array) – Input target time series signal.
- metric (str, optional) – The distance metric to use; one of ‘euclidean’ or ‘pearson’; default is ‘euclidean’.
Returns: dist (array) – Distance of the query sequence to every sub-sequnce in the signal.
Notes
- Computes distances on z-normalized data.
References
[Mueen2014] Abdullah Mueen, Hossein Hamooni, “Trilce Estrada: Time Series Join on Subsequence Correlation”, ICDM 2014: 450-459
-
biosppy.signals.tools.filter_signal(signal=None, ftype='FIR', band='lowpass', order=None, frequency=None, sampling_rate=1000.0, **kwargs)¶ Filter a signal according to the given parameters.
Parameters: - signal (array) – Signal to filter.
- ftype (str) –
- Filter type:
- Finite Impulse Response filter (‘FIR’);
- Butterworth filter (‘butter’);
- Chebyshev filters (‘cheby1’, ‘cheby2’);
- Elliptic filter (‘ellip’);
- Bessel filter (‘bessel’).
- band (str) –
- Band type:
- Low-pass filter (‘lowpass’);
- High-pass filter (‘highpass’);
- Band-pass filter (‘bandpass’);
- Band-stop filter (‘bandstop’).
- order (int) – Order of the filter.
- frequency (int, float, list, array) –
- Cutoff frequencies; format depends on type of band:
- ’lowpass’ or ‘bandpass’: single frequency;
- ’bandpass’ or ‘bandstop’: pair of frequencies.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- **kwargs (dict, optional) – Additional keyword arguments are passed to the underlying scipy.signal function.
Returns: - signal (array) – Filtered signal.
- sampling_rate (float) – Sampling frequency (Hz).
- params (dict) – Filter parameters.
Notes
- Uses a forward-backward filter implementation. Therefore, the combined filter has linear phase.
-
biosppy.signals.tools.find_extrema(signal=None, mode='both')¶ Locate local extrema points in a signal.
Based on Fermat’s Theorem [Ferm].
Parameters: - signal (array) – Input signal.
- mode (str, optional) – Whether to find maxima (‘max’), minima (‘min’), or both (‘both’).
Returns: - extrema (array) – Indices of the extrama points.
- values (array) – Signal values at the extrema points.
References
[Ferm] Wikipedia, “Fermat’s theorem (stationary points)”, https://en.wikipedia.org/wiki/Fermat%27s_theorem_(stationary_points)
-
biosppy.signals.tools.find_intersection(x1=None, y1=None, x2=None, y2=None, alpha=1.5, xtol=1e-06, ytol=1e-06)¶ Find the intersection points between two lines using piecewise polynomial interpolation.
Parameters: - x1 (array) – Array of x-coordinates of the first line.
- y1 (array) – Array of y-coordinates of the first line.
- x2 (array) – Array of x-coordinates of the second line.
- y2 (array) – Array of y-coordinates of the second line.
- alpha (float, optional) – Resolution factor for the x-axis; fraction of total number of x-coordinates.
- xtol (float, optional) – Tolerance for the x-axis.
- ytol (float, optional) – Tolerance for the y-axis.
Returns: - roots (array) – Array of x-coordinates of found intersection points.
- values (array) – Array of y-coordinates of found intersection points.
Notes
- If no intersection is found, returns the closest point.
-
biosppy.signals.tools.finite_difference(signal=None, weights=None)¶ Apply the Finite Difference method to compute derivatives.
Parameters: - signal (array) – Signal to differentiate.
- weights (list, array) – Finite difference weight coefficients.
Returns: - index (array) – Indices from signal for which the derivative was computed.
- derivative (array) – Computed derivative.
Notes
- The method assumes central differences weights.
- The method accounts for the delay introduced by the algorithm.
Raises: ValueError– If the number of weights is not odd.
-
biosppy.signals.tools.get_filter(ftype='FIR', band='lowpass', order=None, frequency=None, sampling_rate=1000.0, **kwargs)¶ Compute digital (FIR or IIR) filter coefficients with the given parameters.
Parameters: - ftype (str) –
- Filter type:
- Finite Impulse Response filter (‘FIR’);
- Butterworth filter (‘butter’);
- Chebyshev filters (‘cheby1’, ‘cheby2’);
- Elliptic filter (‘ellip’);
- Bessel filter (‘bessel’).
- band (str) –
- Band type:
- Low-pass filter (‘lowpass’);
- High-pass filter (‘highpass’);
- Band-pass filter (‘bandpass’);
- Band-stop filter (‘bandstop’).
- order (int) – Order of the filter.
- frequency (int, float, list, array) –
- Cutoff frequencies; format depends on type of band:
- ’lowpass’ or ‘bandpass’: single frequency;
- ’bandpass’ or ‘bandstop’: pair of frequencies.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- **kwargs (dict, optional) – Additional keyword arguments are passed to the underlying scipy.signal function.
Returns: - b (array) – Numerator coefficients.
- a (array) – Denominator coefficients.
- See Also – scipy.signal
- ftype (str) –
-
biosppy.signals.tools.get_heart_rate(beats=None, sampling_rate=1000.0, smooth=False, size=3)¶ Compute instantaneous heart rate from an array of beat indices.
Parameters: - beats (array) – Beat location indices.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- smooth (bool, optional) – If True, perform smoothing on the resulting heart rate.
- size (int, optional) – Size of smoothing window; ignored if smooth is False.
Returns: - index (array) – Heart rate location indices.
- heart_rate (array) – Instantaneous heart rate (bpm).
Notes
- Assumes normal human heart rate to be between 40 and 200 bpm.
-
biosppy.signals.tools.mean_waves(data=None, size=None, step=None)¶ Extract mean samples from a data set.
Parameters: - data (array) – An m by n array of m data samples in an n-dimensional space.
- size (int) – Number of samples to use for each mean sample.
- step (int, optional) – Number of samples to jump, controlling overlap; default is equal to size (no overlap).
Returns: waves (array) – An k by n array of mean samples.
Notes
- Discards trailing samples if they are not enough to satify the size parameter.
Raises: ValueError– If step is an invalid value.ValueError– If there are not enough samples for the given size.
-
biosppy.signals.tools.median_waves(data=None, size=None, step=None)¶ Extract median samples from a data set.
Parameters: - data (array) – An m by n array of m data samples in an n-dimensional space.
- size (int) – Number of samples to use for each median sample.
- step (int, optional) – Number of samples to jump, controlling overlap; default is equal to size (no overlap).
Returns: waves (array) – An k by n array of median samples.
Notes
- Discards trailing samples if they are not enough to satify the size parameter.
Raises: ValueError– If step is an invalid value.ValueError– If there are not enough samples for the given size.
-
biosppy.signals.tools.normalize(signal=None, ddof=1)¶ Normalize a signal to zero mean and unitary standard deviation.
Parameters: - signal (array) – Input signal.
- ddof (int, optional) – Delta degrees of freedom for standard deviation computation; the divisor is N - ddof, where N is the number of elements; default is one.
Returns: signal (array) – Normalized signal.
-
biosppy.signals.tools.pearson_correlation(x=None, y=None)¶ Compute the Pearson Correlation Coefficient bertween two signals.
The coefficient is given by:
![r_{xy} = \frac{E[(X - \mu_X) (Y - \mu_Y)]}{\sigma_X \sigma_Y}](_images/math/38de2c3619a7f8d6109ae1514f75edc00afc2adf.png)
Parameters: - x (array) – First input signal.
- y (array) – Second input signal.
Returns: rxy (float) – Pearson correlation coefficient, ranging between -1 and +1.
Raises: ValueError– If the input signals do not have the same length.
-
biosppy.signals.tools.phase_locking(signal1=None, signal2=None, N=None)¶ Compute the Phase-Locking Factor (PLF) between two signals.
Parameters: - signal1 (array) – First input signal.
- signal2 (array) – Second input signal.
- N (int, optional) – Number of Fourier components.
Returns: plf (float) – The PLF between the two signals.
-
biosppy.signals.tools.power_spectrum(signal=None, sampling_rate=1000.0, pad=None, pow2=False, decibel=True)¶ Compute the power spectrum of a signal (one-sided).
Parameters: - signal (array) – Input signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- pad (int, optional) – Padding for the Fourier Transform (number of zeros added); defaults to no padding..
- pow2 (bool, optional) – If True, rounds the number of points N = len(signal) + pad to the nearest power of 2 greater than N.
- decibel (bool, optional) – If True, returns the power in decibels.
Returns: - freqs (array) – Array of frequencies (Hz) at which the power was computed.
- power (array) – Power spectrum.
-
biosppy.signals.tools.rms_error(x=None, y=None)¶ Compute the Root-Mean-Square Error between two signals.
The error is given by:
![rmse = \sqrt{E[(X - Y)^2]}](_images/math/5b1ba38b9afb4d8cbfffa3061176469a0b6562f7.png)
Parameters: - x (array) – First input signal.
- y (array) – Second input signal.
Returns: rmse (float) – Root-mean-square error.
Raises: ValueError– If the input signals do not have the same length.
-
biosppy.signals.tools.signal_cross_join(signal1=None, signal2=None, size=None, index=None, limit=None)¶ Compute the matrix profile for a similarity join of two time series.
Computes the nearest sub-sequence in signal2 for each sub-sequence in signal1. Implements the algorithm described in [Yeh2016_c].
Parameters: - signal1 (array) – Fisrt input time series signal.
- signal2 (array) – Second input time series signal.
- size (int) – Size of the query sub-sequences.
- index (list, array, optional) – Starting indices for query sub-sequences; the default is to search all sub-sequences.
- limit (int, optional) – Upper limit for the number of query sub-sequences; the default is to search all sub-sequences.
Returns: - matrix_index (array) – Matric profile index.
- matrix_profile (array) – Computed matrix profile (distances).
Notes
- Computes euclidean distances on z-normalized data.
References
[Yeh2016_c] Chin-Chia Michael Yeh, Yan Zhu, Liudmila Ulanova, Nurjahan Begum, Yifei Ding, Hoang Anh Dau, Diego Furtado Silva, Abdullah Mueen, Eamonn Keogh, “Matrix Profile I: All Pairs Similarity Joins for Time Series: A Unifying View that Includes Motifs, Discords and Shapelets”, IEEE ICDM 2016
-
biosppy.signals.tools.signal_self_join(signal=None, size=None, index=None, limit=None)¶ Compute the matrix profile for a self-similarity join of a time series.
Implements the algorithm described in [Yeh2016_b].
Parameters: - signal (array) – Input target time series signal.
- size (int) – Size of the query sub-sequences.
- index (list, array, optional) – Starting indices for query sub-sequences; the default is to search all sub-sequences.
- limit (int, optional) – Upper limit for the number of query sub-sequences; the default is to search all sub-sequences.
Returns: - matrix_index (array) – Matric profile index.
- matrix_profile (array) – Computed matrix profile (distances).
Notes
- Computes euclidean distances on z-normalized data.
References
[Yeh2016_b] Chin-Chia Michael Yeh, Yan Zhu, Liudmila Ulanova, Nurjahan Begum, Yifei Ding, Hoang Anh Dau, Diego Furtado Silva, Abdullah Mueen, Eamonn Keogh, “Matrix Profile I: All Pairs Similarity Joins for Time Series: A Unifying View that Includes Motifs, Discords and Shapelets”, IEEE ICDM 2016
-
biosppy.signals.tools.signal_stats(signal=None)¶ Compute various metrics describing the signal.
Parameters: signal (array) – Input signal. Returns: - mean (float) – Mean of the signal.
- median (float) – Median of the signal.
- max (float) – Maximum signal amplitude.
- var (float) – Signal variance (unbiased).
- std_dev (float) – Standard signal deviation (unbiased).
- abs_dev (float) – Absolute signal deviation.
- kurtosis (float) – Signal kurtosis (unbiased).
- skew (float) – Signal skewness (unbiased).
-
biosppy.signals.tools.smoother(signal=None, kernel='boxzen', size=10, mirror=True, **kwargs)¶ Smooth a signal using an N-point moving average [MAvg] filter.
This implementation uses the convolution of a filter kernel with the input signal to compute the smoothed signal [Smit97].
Availabel kernels: median, boxzen, boxcar, triang, blackman, hamming, hann, bartlett, flattop, parzen, bohman, blackmanharris, nuttall, barthann, kaiser (needs beta), gaussian (needs std), general_gaussian (needs power, width), slepian (needs width), chebwin (needs attenuation).
Parameters: - signal (array) – Signal to smooth.
- kernel (str, array, optional) – Type of kernel to use; if array, use directly as the kernel.
- size (int, optional) – Size of the kernel; ignored if kernel is an array.
- mirror (bool, optional) – If True, signal edges are extended to avoid boundary effects.
- **kwargs (dict, optional) – Additional keyword arguments are passed to the underlying scipy.signal.windows function.
Returns: - signal (array) – Smoothed signal.
- params (dict) – Smoother parameters.
Notes
- When the kernel is ‘median’, mirror is ignored.
References
[MAvg] Wikipedia, “Moving Average”, http://en.wikipedia.org/wiki/Moving_average [Smit97] S. W. Smith, “Moving Average Filters - Implementation by Convolution”, http://www.dspguide.com/ch15/1.htm, 1997
-
biosppy.signals.tools.synchronize(x=None, y=None, detrend=True)¶ Align two signals based on cross-correlation.
Parameters: - x (array) – First input signal.
- y (array) – Second input signal.
- detrend (bool, optional) – If True, remove signal means before computation.
Returns: - delay (int) – Delay (number of samples) of ‘x’ in relation to ‘y’; if ‘delay’ < 0 , ‘x’ is ahead in relation to ‘y’; if ‘delay’ > 0 , ‘x’ is delayed in relation to ‘y’.
- corr (float) – Value of maximum correlation.
- synch_x (array) – Biggest possible portion of ‘x’ in synchronization.
- synch_y (array) – Biggest possible portion of ‘y’ in synchronization.
-
biosppy.signals.tools.welch_spectrum(signal=None, sampling_rate=1000.0, size=None, overlap=None, window='hanning', window_kwargs=None, pad=None, decibel=True)¶ Compute the power spectrum of a signal using Welch’s method (one-sided).
Parameters: - signal (array) – Input signal.
- sampling_rate (int, float, optional) – Sampling frequency (Hz).
- size (int, optional) – Number of points in each Welch segment; defaults to the equivalent of 1 second; ignored when ‘window’ is an array.
- overlap (int, optional) – Number of points to overlap between segments; defaults to size / 2.
- window (str, array, optional) – Type of window to use.
- window_kwargs (dict, optional) – Additional keyword arguments to pass on window creation; ignored if ‘window’ is an array.
- pad (int, optional) – Padding for the Fourier Transform (number of zeros added); defaults to no padding.
- decibel (bool, optional) – If True, returns the power in decibels.
Returns: - freqs (array) – Array of frequencies (Hz) at which the power was computed.
- power (array) – Power spectrum.
Notes
- Detrends each Welch segment by removing the mean.
-
biosppy.signals.tools.windower(signal=None, size=None, step=None, fcn=None, fcn_kwargs=None, kernel='boxcar', kernel_kwargs=None)¶ Apply a function to a signal in sequential windows, with optional overlap.
Availabel window kernels: boxcar, triang, blackman, hamming, hann, bartlett, flattop, parzen, bohman, blackmanharris, nuttall, barthann, kaiser (needs beta), gaussian (needs std), general_gaussian (needs power, width), slepian (needs width), chebwin (needs attenuation).
Parameters: - signal (array) – Input signal.
- size (int) – Size of the signal window.
- step (int, optional) – Size of window shift; if None, there is no overlap.
- fcn (callable) – Function to apply to each window.
- fcn_kwargs (dict, optional) – Additional keyword arguments to pass to ‘fcn’.
- kernel (str, array, optional) – Type of kernel to use; if array, use directly as the kernel.
- kernel_kwargs (dict, optional) – Additional keyword arguments to pass on window creation; ignored if ‘kernel’ is an array.
Returns: - index (array) – Indices characterizing window locations (start of the window).
- values (array) – Concatenated output of calling ‘fcn’ on each window.
-
biosppy.signals.tools.zero_cross(signal=None, detrend=False)¶ Locate the indices where the signal crosses zero.
Parameters: - signal (array) – Input signal.
- detrend (bool, optional) – If True, remove signal mean before computation.
Returns: zeros (array) – Indices of zero crossings.
Notes
- When the signal crosses zero between samples, the first index is returned.